Transition networks with python
First the imports in ipython notebook:
%pylab inline
import skimage.morphology as morphology
import sklearn.cluster
import graphviz
import os
import PIL
import matplotlib.gridspec as gridspecIdentifying metastable states
Understanding a system’s conformational dynamics can be summed up into two steps:
-
identifying the long lived, or metastable, states visited by the system
-
determining the rates of transitioning between these states.
I decided to reproduce in python the results obtained in this publication:
Transition networks for modeling the kinetics of conformational change in macromolecules
by Frank Noé, Stefan Fischer
DOI: 10.1016/j.sbi.2008.01.008
abstract:
The kinetics and thermodynamics of complex transitions in biomolecules can be modeled in terms of a network of transitions between the relevant conformational substates. Such a transition network, which overcomes the fundamental limitations of reaction-coordinate-based methods, can be constructed either based on the features of the energy landscape, or from molecular dynamics simulations. Energy-landscape-based networks are generated with the aid of automated path-optimization methods, and, using graph-theoretical adaptive methods, can now be constructed for large molecules such as proteins. Dynamics- based networks, also called Markov State Models, can be interpreted and adaptively improved using statistical concepts, such as the mean first passage time, reactive flux and sampling error analysis. This makes transition networks powerful tools for understanding large-scale conformational changes.
The 1D energy of the system
x = linspace(0,100,7)
y = [5, 1.5, 4, 1, 2, 1.5,5]
plot(x,y,'o')
z = np.polyfit(x, y, 6)
E = np.poly1d(z)
x_lin = linspace(0, 100, 100)
plot(x_lin,E(x_lin), linewidth=2)
ylabel('Energy ($k_{B}T$)', fontsize=18)
xlabel('discrete coordinates', fontsize=18)
grid()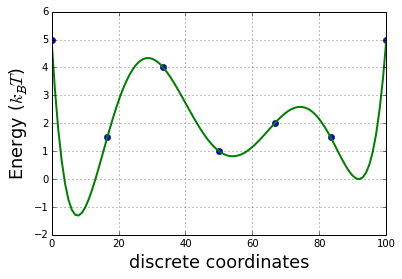
This energy plot is sample with a basic Markov Chain Monte Carlo (MCMC)
def montecarlo(potential=E, nstep=1000):
p = lambda x: exp(-x)
pos_prev = random.randint(0,100)
traj = []
for i in range(nstep):
pos = pos_prev + randint(0, 2)*2 -1
while not (0 <= pos < 100):
pos = pos_prev + randint(0, 2)*2 -1
delta = potential(pos) - potential(pos_prev)
if delta > 0:
#print p(delta)
if p(delta) < random.uniform():
pos = pos_prev
else:
pos_prev = pos
else:
pos_prev = pos
#print pos, pos_prev, delta, potential(pos), potential(pos_prev)
traj.append(pos)
return asarray(traj)1000000 steps of MCMC and we compute the transition matrix
traj = montecarlo(nstep=1000000)Here the distribution obtained with th MCMC which fit quite well the energy plot
tmp = hist(traj, 100, normed=True)
ylabel('$\pi$', fontsize=18)
xlabel('discrete coordinates', fontsize=18)
grid()
def get_transition_matrix(traj, lag=1):
T = zeros((100, 100))
density = zeros(100, dtype=int)
for e in zip(traj, traj[lag:]):
i,j = e
T[i,j] += 1
density[i] += 1
T /= density
return nan_to_num(T)
T = get_transition_matrix(traj, lag=200)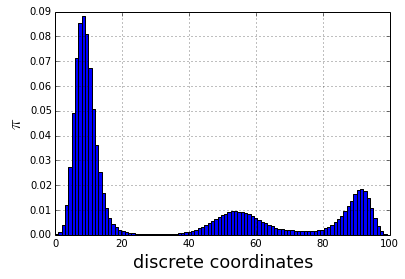
The plot of the transition matrix
imshow(T, interpolation='nearest', cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())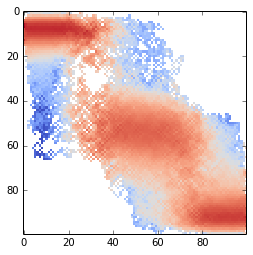
The transition matrix is then diagonalized
w,v = linalg.eig(T)
w = real(w)
v = real(v)
v = v[:,w.argsort()[::-1]]
w = w[w.argsort()[::-1]]Just a simple watershed on the energy to identify energy basins based on the energy plot
structelem = numpy.ones((3), dtype='bool')
labels = zeros(100)
for i,e in enumerate(x[1:6:2]):
labels[int(e)] = i+1
x_lin = linspace(0, 100, 100)
labels_full = morphology.watershed(E(x_lin), labels, connectivity=structelem)A function to substitute values in an array
def substitute(d, arr):
"""
d: {0: 3, 1: 1, 2: 2} for example: values to substitute
"""
new_arr = copy.deepcopy(arr)
for i, j in d.iteritems():
new_arr[arr==i] = j
return new_arrThe projection of the transition matrix onto the second and third eigen vector
proj = dot(transpose(T),v[:,1:3])
# create k-meansclustering estimators
kmeans = sklearn.cluster.KMeans(n_clusters=3)
kmeans.fit(proj)
mgrid = asarray(meshgrid(linspace(min(proj[:,0]),max(proj[:,0]),100),linspace(min(proj[:,1]),max(proj[:,1]),100))).T
kmeans_partition = kmeans.predict(mgrid.reshape(100*100,2)).reshape(100,100)
d = {}
for i in unique(kmeans.labels_):
c,b = histogram(labels_full[kmeans.labels_==i])
d[i] = int(b[argmax(c)])
print d
imshow(kmeans_partition[:,::-1].T, interpolation='none', extent=[min(proj[:,0]),max(proj[:,0]),min(proj[:,1]),max(proj[:,1])])
colorbar()
scatter(proj[:,0], proj[:,1],c=labels_full, s=64)
xlabel('second eigenvector')
ylabel('third eigenvector')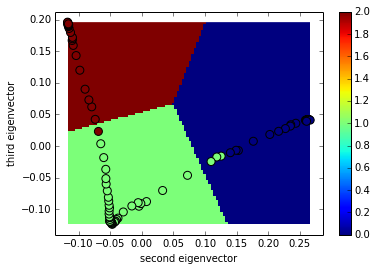
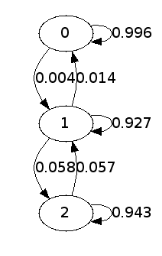
The code below identifies the three metastable states and compute the Markov chain model
pi, bins = histogram(traj, normed=True, bins=100)
plot(pi)
xlabel('discrete coordinates', fontsize = 18)
ylabel('$\pi$', fontsize=18)
def meta_transition(c1,c2):
sel1 = kmeans.labels_ == c1
sel2 = kmeans.labels_ == c2
ind1 = where(sel1)[0]
ind2 = where(sel2)[0]
num = 0
for i in ind1:
for j in ind2:
num+=pi[i] * T[j,i]
trans = num/pi[sel1].sum()
return trans
gdot = graphviz.Digraph()
for i in unique(kmeans.labels_):
gdot.node('%d'%i)
for i in unique(kmeans.labels_):
for j in unique(kmeans.labels_):
weight = meta_transition(i,j)
if weight > 0:
gdot.edge('%d'%i, '%d'%j, label='%.3f'%weight)
dotfn = 'graph.dot'
gdot.render(filename=dotfn)
def plotdotgraph(dotfn):
os.system('dot -T png -o %s.png %s'%(dotfn,dotfn))
img = PIL.Image.open('%s.png'%dotfn)
imshow(img)
axis('off')
figure()
tmp = plotdotgraph(dotfn)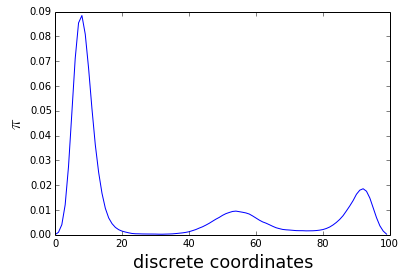
And here the full story in one plot:
rcParams['figure.figsize'] = 18,10
gs = gridspec.GridSpec(4,3, height_ratios=(1,1,1,3))
gs.update(hspace=.5)
subplot(gs[:3,0])
x_lin = linspace(0, 100, 100)
for i in range(3):
plot(x_lin[labels_full==i+1],E(x_lin)[labels_full==i+1], linewidth=2)
ylabel('Energy ($k_{B}T$)', fontsize=18)
xlabel('discrete coordinates', fontsize=18)
grid()
subplot(gs[:3,1])
imshow(T, interpolation='nearest', cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
for i in range(3):
subplot(gs[i,2])
for j in range(3):
plot(x_lin[labels_full==j+1],-v[:,i][labels_full==j+1], linewidth=2)
grid()
xlabel('discrete coordinates', fontsize=18)
subplot(gs[3,0])
plot(w[:10], 'o--')
xlabel('eigenvalue index', fontsize=18)
ylabel('eigenvalue', fontsize=18)
grid()
subplot(gs[3,1])
imshow(kmeans_partition[:,::-1].T, interpolation='none', extent=[min(proj[:,0]),max(proj[:,0]),min(proj[:,1]),max(proj[:,1])],
alpha=.5)
scatter(proj[:,0], proj[:,1],c=labels_full, s=64)
xlabel('second eigenvector', fontsize=18)
ylabel('third eigenvector', fontsize=18)
grid()
subplot(gs[3,2])
plotdotgraph(dotfn)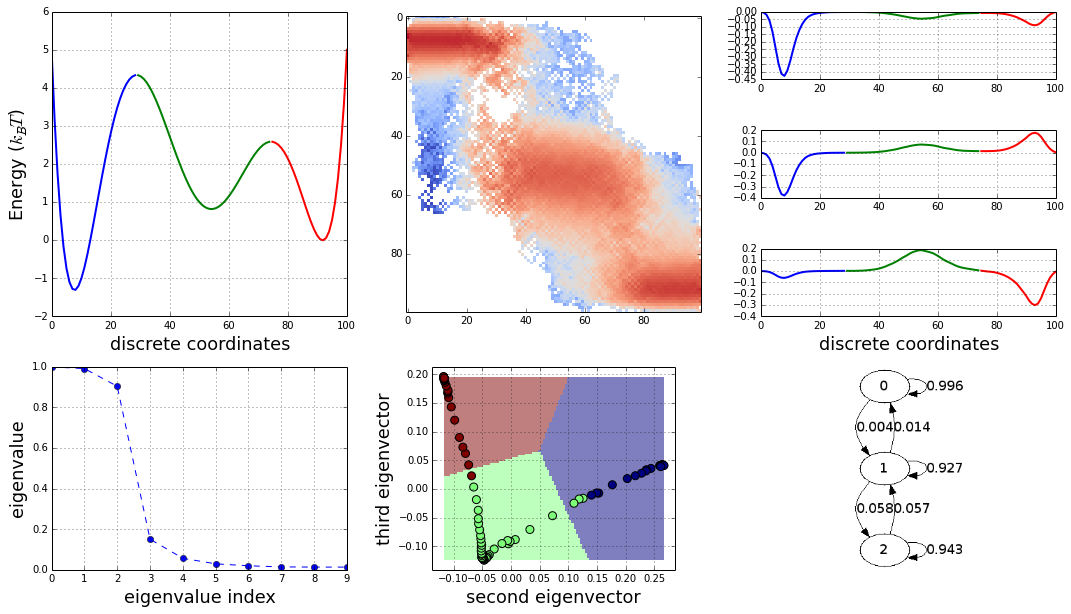
The construction of a transition network. (a) Sample potential, defined over a one-dimensional coordinate that is discretized into 100 microstates. It has three metastable basins (0 (blue), 1 (green), and 2 (red)). (b) Transition matrix T for a Markov lagtime of 200 steps. The transition probability was obtained from a Metropolis Monte Carlo, jumping each step only to the current or adjacent microstates. T exhibits three clusters corresponding to the metastable states. (c) Left eigenvectors of T indicating the transition modes among microstates. The first eigenvector gives the stationary distribution. The sign structure of the second eigenvector partitions the state space into two metastable states, blue and red. The sign structure of the third eigenvector further splits green and red, obtaining three metastable states. (d) The eigenvalue spectrum of T. The clear gaps after 2 and 3 eigenvalues indicate how many states are metastable. (e) Coordinates of the 100 microstates projected onto the second and third right eigenvectors of T. Metastable states are identified by clustering the microstates in this eigenspace. (f) Transition network between the 3 metastable states A, B and C
The caption is adapted from: Transition networks for modeling the kinetics of conformational change in macromolecules
If you want to ask me a question or leave me a message add @bougui505 in your comment.