Simple Markov chain Monte Carlo (MCMC) algorithm in python
I present here a simple Markov chain Monte Carlo python implementation to sample a 2D surface of potential.
%pylab inlineFirst I define a Müller potential from a piece of code I found on gist.
def muller_potential(x, y, use_numpy=False):
"""Muller potential
Parameters
----------
x : {float, np.ndarray, or theano symbolic variable}
X coordinate. If you supply an array, x and y need to be the same shape,
and the potential will be calculated at each (x,y pair)
y : {float, np.ndarray, or theano symbolic variable}
Y coordinate. If you supply an array, x and y need to be the same shape,
and the potential will be calculated at each (x,y pair)
Returns
-------
potential : {float, np.ndarray, or theano symbolic variable}
Potential energy. Will be the same shape as the inputs, x and y.
Reference
---------
Code adapted from https://cims.nyu.edu/~eve2/ztsMueller.m
"""
aa = [-1, -1, -6.5, 0.7]
bb = [0, 0, 11, 0.6]
cc = [-10, -10, -6.5, 0.7]
AA = [-200, -100, -170, 15]
XX = [1, 0, -0.5, -1]
YY = [0, 0.5, 1.5, 1]
# use symbolic algebra if you supply symbolic quantities
exp = np.exp
value = 0
for j in range(0, 4):
if use_numpy:
value += AA[j] * numpy.exp(aa[j] * (x - XX[j])**2 + bb[j] * (x - XX[j]) * (y - YY[j]) + cc[j] * (y - YY[j])**2)
else: # use sympy
value += AA[j] * sympy.exp(aa[j] * (x - XX[j])**2 + bb[j] * (x - XX[j]) * (y - YY[j]) + cc[j] * (y - YY[j])**2)
return valueWhich looks like:
minx=-1.5
maxx=1.2
miny=-0.2
maxy=2
ax=None
grid_width = max(maxx-minx, maxy-miny) / 200.0
xx, yy = np.mgrid[minx : maxx : grid_width, miny : maxy : grid_width]
V = muller_potential(xx, yy, use_numpy=True)
contourf(xx, yy, ma.masked_array(V.clip(max=200), V>inf), 40)
tmp = colorbar()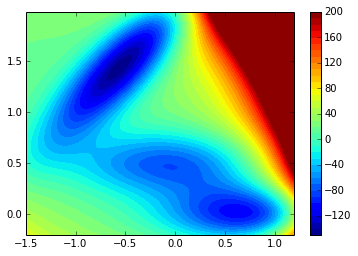
The code for MCMC sampling is quite short:
def montecarlo(potential=V, nstep=1000, beta=1, markov=True):
p = lambda x: exp(-beta*x)
nx,ny = potential.shape
pos_prev = (np.random.randint(0,nx), np.random.randint(0,ny))
traj = []
for i in range(nstep):
if markov:
pos = (pos_prev + asarray([random.choice([-1,0,1]), random.choice([-1,0,1])]))%(nx,ny)
else:
pos = (np.random.randint(0,nx), np.random.randint(0,ny))
pos = tuple(pos)
pos_prev = tuple(pos_prev)
delta = potential[pos] - potential[pos_prev]
if delta > 0:
#print p(delta)
if p(delta) < np.random.uniform():
pos = pos_prev
else:
pos_prev = pos
else:
pos_prev = pos
traj.append(pos)
return asarray(traj)And takes less than 1 minute for 1 000 000 steps of monte carlo:
nstep=1000000
traj = montecarlo(nstep=nstep, beta=0.125, markov=False)
density = zeros_like(V)
for pos in traj:
pos = tuple(pos)
density[pos] += 1Now we can plot the distribution obtained onto the surface of the potential sampled:
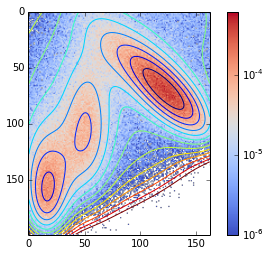
contour(V.clip(max=200), 15)
imshow(density / nstep, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
tmp = colorbar()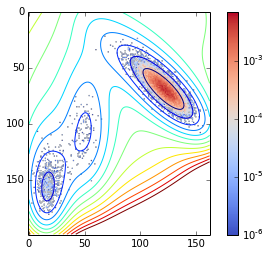
And if you increase :
nstep=1000000
traj = montecarlo(nstep=nstep, beta=0.025, markov=False)
density = zeros_like(V)
for pos in traj:
pos = tuple(pos)
density[pos] += 1You sample more:
contour(V.clip(max=200), 15)
imshow(density / nstep, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
tmp = colorbar()