Metadynamics on a 2D surface potential with python
First, the python import in the ipython notebook:
%pylab inline
import matplotlib.gridspec as gridspec
import scipy.spatial.distance
from skimage.feature import peak_local_max
import scipy.ndimage.filters
import copy
from matplotlib import animation
from JSAnimation import IPython_display
from JSAnimation.IPython_display import display_animationPopulating the interactive namespace from numpy and matplotlibAs usual, we define the Müller potential as the sampled potential:
def muller_potential(x, y):
"""Muller potential
Parameters
----------
x : {float, np.ndarray, or theano symbolic variable}
X coordinate. If you supply an array, x and y need to be the same shape,
and the potential will be calculated at each (x,y pair)
y : {float, np.ndarray, or theano symbolic variable}
Y coordinate. If you supply an array, x and y need to be the same shape,
and the potential will be calculated at each (x,y pair)
Returns
-------
potential : {float, np.ndarray, or theano symbolic variable}
Potential energy. Will be the same shape as the inputs, x and y.
Reference
---------
Code adapted from https://cims.nyu.edu/~eve2/ztsMueller.m
"""
aa = [-1, -1, -6.5, 0.7]
bb = [0, 0, 11, 0.6]
cc = [-10, -10, -6.5, 0.7]
AA = [-200, -100, -170, 15]
XX = [1, 0, -0.5, -1]
YY = [0, 0.5, 1.5, 1]
# use symbolic algebra if you supply symbolic quantities
exp = np.exp
value = 0
for j in range(0, 4):
value += AA[j] * numpy.exp(aa[j] * (x - XX[j])**2 + bb[j] * (x - XX[j]) * (y - YY[j]) + cc[j] * (y - YY[j])**2)
return value2D plot of the sampled potential:
minx=-1.5
maxx=1.2
miny=-0.2
maxy=2
ax=None
grid_width = max(maxx-minx, maxy-miny) / 200.0
xx, yy = np.mgrid[minx : maxx : grid_width, miny : maxy : grid_width]
V = muller_potential(xx, yy)
contourf(xx, yy, V.clip(max=200), 40)
colorbar()<matplotlib.colorbar.Colorbar instance at 0xd54d3f8>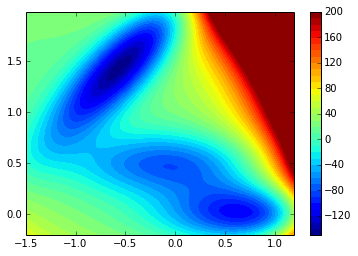
Now, we search for the 3 local minima of the potential defined:
print xx.shape
local_minima_grid = peak_local_max(-V)
local_minima = []
for e in local_minima_grid:
local_minima.append([xx[tuple(e)], yy[tuple(e)]])
local_minima = asarray(local_minima)(200, 163)contourf(xx, yy, V.clip(max=200), 40)
colorbar()
scatter(local_minima[:,0], local_minima[:,1], c='r')<matplotlib.collections.PathCollection at 0xd96d910>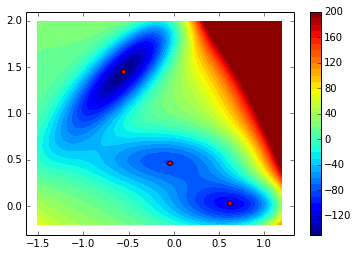
And now the standard MCMC sampler:
def montecarlo(start=None,potential=V, nstep=1000, beta=1, markov=True):
p = lambda x: exp(-beta*x)
nx,ny = potential.shape
if start == None:
pos_prev = (np.random.randint(0,nx), np.random.randint(0,ny))
else:
pos_prev = start
traj = []
for i in range(nstep):
if markov:
pos = (pos_prev + asarray([random.choice([-1,0,1]), random.choice([-1,0,1])]))%(nx,ny)
else:
pos = (np.random.randint(0,nx), np.random.randint(0,ny))
pos = tuple(pos)
pos_prev = tuple(pos_prev)
delta = potential[pos] - potential[pos_prev]
if delta > 0:
#print p(delta)
if p(delta) < np.random.uniform():
pos = pos_prev
else:
pos_prev = pos
else:
pos_prev = pos
traj.append(pos)
return asarray(traj)The sampling:
nstep=100000
traj = montecarlo(start = local_minima_grid[2], nstep=nstep, beta=1, markov=True)And the distribution obtained:
def get_density(traj):
density = zeros_like(V)
for pos in traj:
pos = tuple(pos)
density[pos] += 1
return density
density = get_density(traj)contour(V.clip(max=200), 15)
imshow(density / nstep, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
tmp = colorbar()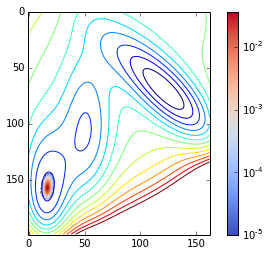
Now, we define a Gaussian for the sampling by metadynamics:
def Gaussian(x,y,x0,y0,w=1,sigma=1):
Z = w*numpy.exp(- ((x-x0)**2 + (y-y0)**2) / (2*sigma**2))
return ZAnd the MCMC sampler is modified to add a Gaussian at each point visited during the sampling:
def metamontecarlo(start = None, potential=V, nstep=1000, beta=1, markov=True):
potential_flooding = []
p = lambda x: exp(-beta*x)
nx,ny = potential.shape
if start == None:
pos_prev = (np.random.randint(0,nx), np.random.randint(0,ny))
else:
pos_prev = start
traj = []
for i in range(nstep):
if markov:
pos = (pos_prev + asarray([random.choice([-1,0,1]), random.choice([-1,0,1])]))%(nx,ny)
else:
pos = (np.random.randint(0,nx), np.random.randint(0,ny))
pos = tuple(pos)
pos_prev = tuple(pos_prev)
delta = potential[pos] - potential[pos_prev]
if delta > 0:
#print p(delta)
if p(delta) < np.random.uniform():
pos = pos_prev
else:
pos_prev = pos
else:
pos_prev = pos
potential += Gaussian(xx,yy, xx[pos],yy[pos], w=0.01, sigma=10*grid_width)
#potential_k = copy.deepcopy(potential)
#potential_flooding.append(potential_k)
traj.append(pos)
return asarray(traj)#, asarray(potential_flooding)Now we sample with the same parameters beta and nstep but with the ‘Gaussian
flooding’
nstep=100000
potential = copy.deepcopy(V)
traj = metamontecarlo(start = local_minima_grid[2], potential=potential, nstep=nstep, beta=1, markov=True)
density = zeros_like(V)
for pos in traj:
pos = tuple(pos)
density[pos] += 1save('traj_meta', traj)traj_meta = load('traj_meta.npy')Now we’ll use the JSAnimation module to generate an animation of the sampling:
fig = plt.figure()
axis('off')
potential_flood = copy.deepcopy(V)
nx,ny = potential.shape
ims = []
for k in range(nstep):
pos = tuple(traj_meta[k])
potential_flood += Gaussian(xx,yy, xx[pos],yy[pos], w=0.01, sigma=10*grid_width)
if k%100 == 0:
potential_k = copy.deepcopy(potential_flood)
for i in range(3):
for j in range(3):
u, v = pos[0]+i, pos[1]+j
if u < nx and v < ny:
potential_k[u,v] = 200
im = plt.imshow(potential_k, interpolation='nearest', vmin = V.min(), vmax=200)
ims.append([im])
animation.ArtistAnimation(fig, ims, interval=50, blit=True,
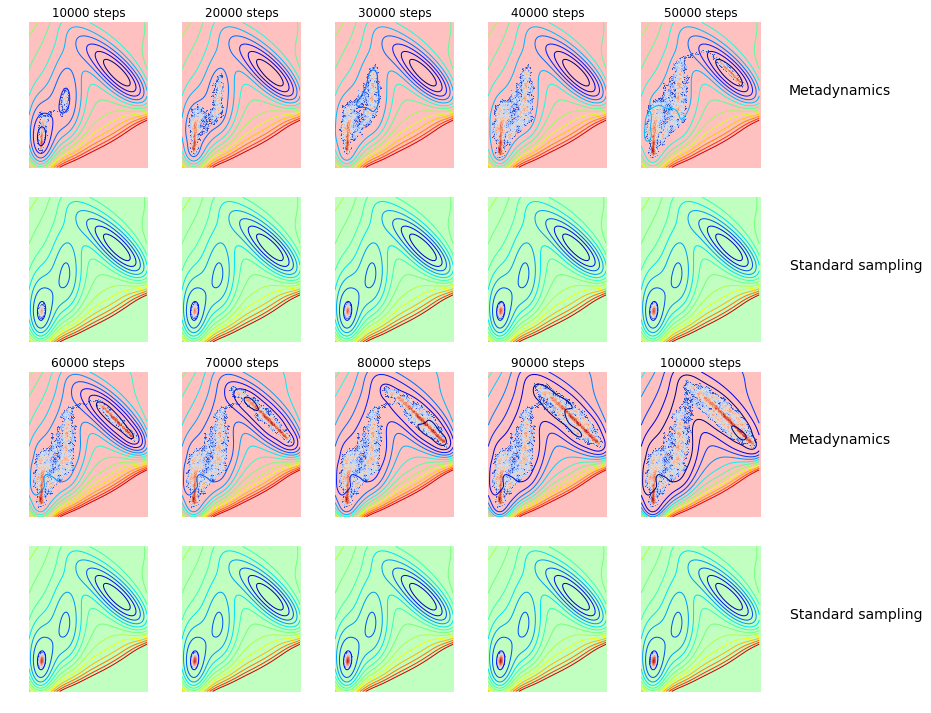
repeat_delay=1000)Below, just some snapshots of the metadynamics and standard sampling for comparison:
max_density_meta = get_density(traj_meta).max()
max_density = get_density(traj).max()potential_flood = copy.deepcopy(V)
nx,ny = potential.shape
potential_flooding = []
for k in range(nstep):
pos = tuple(traj_meta[k])
potential_flood += Gaussian(xx,yy, xx[pos],yy[pos], w=0.01, sigma=10*grid_width)
if k%10000 == 0:
potential_k = copy.deepcopy(potential_flood)
potential_flooding.append(potential_k)print len(potential_flooding)10rcParams['figure.figsize'] = 16,12
gs=gridspec.GridSpec(4,6)
bg1 = zeros((nx,ny,3))
bg1[:,:,0] = 1
bg2 = zeros((nx,ny,3))
bg2[:,:,1] = 1
u=0
v=0
trajs = []
trajs_meta= []
delta = 10000
for i,c in enumerate(range(0,nstep,delta)):
trajs.append(traj[i*delta:(i+1)*delta])
subtraj = asarray(trajs).reshape((i+1)*delta,2)
sub_density = get_density(subtraj)
subplot(gs[u+1,v])
imshow(bg2, alpha=.25)
contour(V.clip(max=200), 15, vmin=V.min(), vmax=200)
imshow(sub_density/max_density, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm(), interpolation='nearest', vmax=1)
axis('off')
#starts = starts_list[i]
subplot(gs[u,v])
imshow(bg1, alpha=.25)
contour(potential_flooding[i].clip(max=200), 15, vmax=200)
trajs_meta.append(traj_meta[i*delta:(i+1)*delta])
subtraj_meta = asarray(trajs_meta).reshape((i+1)*delta,2)
sub_density_meta = get_density(subtraj_meta)
imshow(sub_density_meta/max_density_meta, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm(), interpolation='nearest', vmax=1)
title('%d steps'%((i+1)*delta))
axis('off')
v+=1
if v == 5:
subplot(gs[u,v])
text(0,0.5,'Metadynamics', fontsize=14)
axis('off')
subplot(gs[u+1,v])
text(0,0.5,'Standard sampling', fontsize=14)
axis('off')
v = 0
u += 2
savefig('metadynamics.png', dpi=300, bbox_inches='tight' )