WHAM: The Weighted Histogram Analysis Method
First of all, a good introduction to the WHAM method can be found here or here for a local copy.
Sometime, it’s necessary to determine properties from combined simulations under different conditions. The WHAM method allows to unbias the distribution of the combined simulations.
Here, we present the result on a 1D toy model sampled with a Markov Chain Monte Carlo (MCMC)
Here the import in the ipython notebook:
%pylab inline
import matplotlib.gridspec as gridspec
import scipy.optimize as optimThe 1D potential is constructed with a 1D polynomial fit of some points
x = linspace(0,100,7)
y = [5, 1.5, 4, 1, 2, 1.5,5]
plot(x,y,'o')
z = np.polyfit(x, y, 6)
E = np.poly1d(z)
x_lin = linspace(0, 100, 100)
plot(x_lin,E(x_lin), linewidth=2)
ylabel('Potential', fontsize=18)
xlabel('discrete coordinates', fontsize=18)
grid()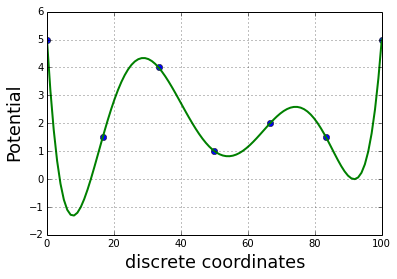
This potential is then sampled using a MCMC
def montecarlo(potential=E, nstep=1000, beta=1):
p = lambda x: exp(-beta*x)
pos_prev = random.randint(0,100)
traj = []
for i in range(nstep):
pos = pos_prev + randint(0, 2)*2 -1
while not (0 <= pos < 100):
pos = pos_prev + randint(0, 2)*2 -1
delta = potential(pos) - potential(pos_prev)
if delta > 0:
#print p(delta)
if p(delta) < random.uniform():
pos = pos_prev
else:
pos_prev = pos
else:
pos_prev = pos
#print pos, pos_prev, delta, potential(pos), potential(pos_prev)
traj.append(pos)
return asarray(traj)We used six different temperature (\(T=\beta^{-1}\)):
-
\(\beta=1.00\)
-
\(\beta=0.85\)
-
\(\beta=0.70\)
-
\(\beta=0.55\)
-
\(\beta=0.40\)
-
\(\beta=0.25\)
beta_list = linspace(1,0.25,6)
traj_list = []
for beta in beta_list:
traj_list.append(montecarlo(nstep=100000, beta=beta))And then we plot the distributions obtained from the 6 MCMC:
rcParams['figure.figsize'] = 16,8
gs = gridspec.GridSpec(2,3)
i,j = 0,0
for c,traj in enumerate(traj_list):
subplot(gs[i,j])
title(r'$\beta$ = %.2f'%beta_list[c], fontsize=18)
if j == 0:
ylabel('$\pi$', fontsize=18)
if i == 1:
xlabel('discrete coordinates', fontsize=18)
grid()
tmp = hist(traj, 100, normed=True)
twinx()
plot(x_lin,E(x_lin), linewidth=2, color='r')
if j == 2:
ylabel('Potential', fontsize=18)
j += 1
if j == 3:
j = 0
i += 1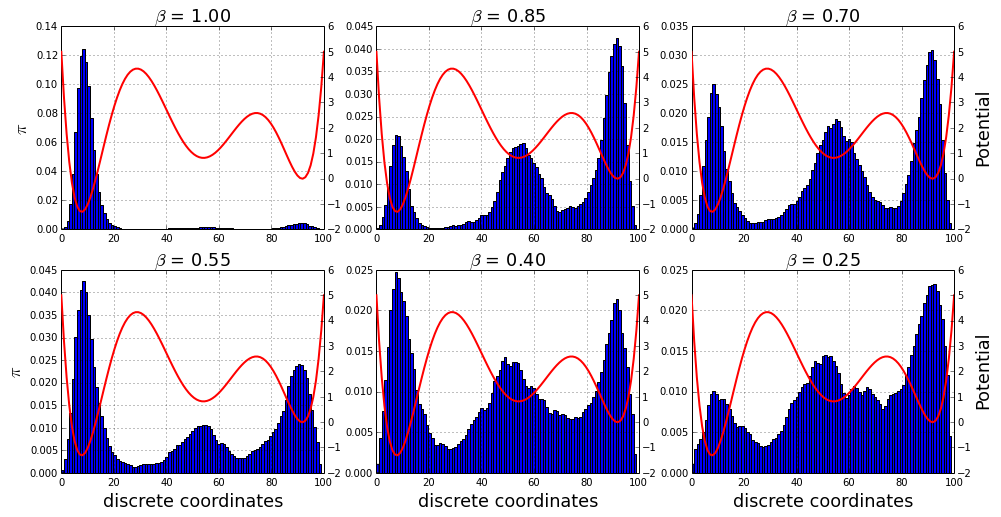
We can look at the distribution of the six MCMC simulations merged together:
rcParams['figure.figsize'] = 16/3,8/2
traj_combined = asarray(traj_list).flatten()
tmp = hist(traj_combined, 100, normed=True)
xlabel('discrete coordinates', fontsize=18)
ylabel('$\pi$', fontsize=18)
twinx()
plot(x_lin,E(x_lin), linewidth=2, color='r')
ylabel('Potential', fontsize=18)
tmp = title('Combined trajectory', fontsize=18)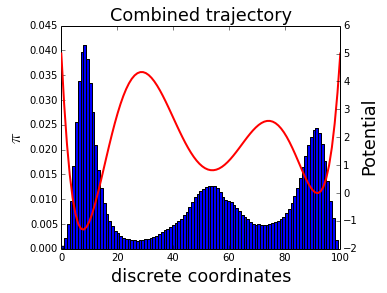
This distribution is biased as we run the MCMC at 6 different temperatures. The aim of WHAM is to unbias the distribution. We assume that , the biased probability of bin j in the i-th simulation, is related to , the unbiased probability of bin j via:
where is the biasing factor and is a normalizing constant chosen such that :
For the temperature biasing:
We assume that we want to compute the potential of mean force (PMF) at \(\beta=1\).
We can compute \(c_{ij}\):
n_sim = len(beta_list) # number of simulations
n_bin = len(x_lin) # number of bins
c = zeros((n_sim, n_bin))
beta0 = 1 # the beta of the unbias distribution we want to compute
for i,beta in enumerate(beta_list):
for j,e in enumerate(E(x_lin)):
c[i,j] = exp(-(beta-beta0)*e)Just for fun, the visualisation of c:
rcParams['figure.figsize'] = 16,5
imshow(c,cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
tmp = colorbar()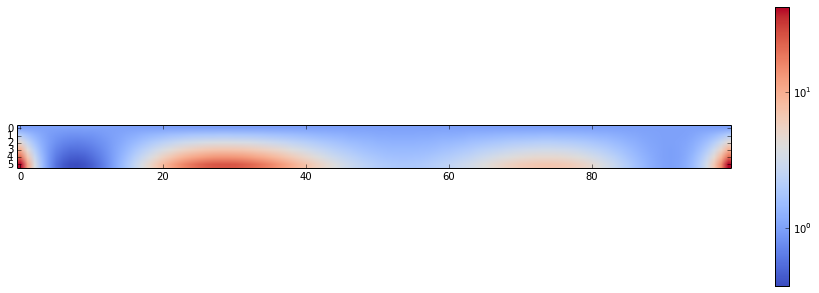
An optimal estimate of \(p_{j}^{o}\) is given by:
with the number of bin count for simulation i and bin j and the total number of sample generated by the i-th simulation.
and, as said before:
S is the total number of simulations (here \(S=6\)) and M is the total number of bins (here \(M=100\)).
We can easily compute \(n_{ij}\;\forall\;(i,j)\)
n = zeros((n_sim, n_bin))
for i,traj in enumerate(traj_list):
n[i] = histogram(traj, bins=100)[0]A representation of n:
matshow(n, cmap=cm.coolwarm, norm=matplotlib.colors.LogNorm())
tmp = colorbar()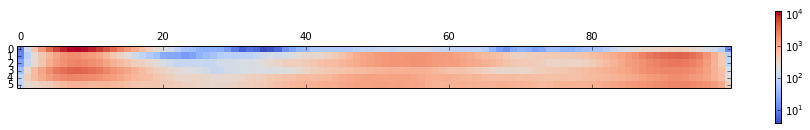
Now we use fsolve to solve the WHAM equations described below:
p0_guess = ones(n_bin, float)
f_guess = ones(n_sim, float)
N = asarray([traj.shape[0] for traj in traj_list])
def residual(P):
p0,f = P[:n_bin],P[n_bin:]
#print p0.shape, f.shape
res1 = p0 - n.sum(axis=0)/(N*f*c.T).T.sum(axis=0)
res2 = f - 1/((c*p0).sum(axis=1))
return concatenate((res1, res2))
#residual(concatenate((p0_guess,f_guess)))
P = optim.fsolve(residual, concatenate((p0_guess, f_guess)))
#print P.x
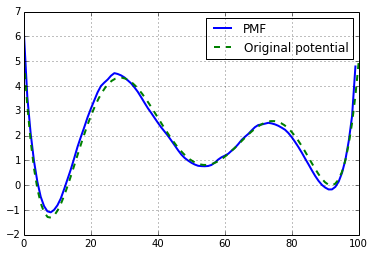
p0,f = P[:n_bin],P[n_bin:]And here the result; the unbias distribution for \(\beta=1\)
plot(p0)
xlabel('discrete coordinates', fontsize=18)
tmp = ylabel('$\pi$', fontsize=18)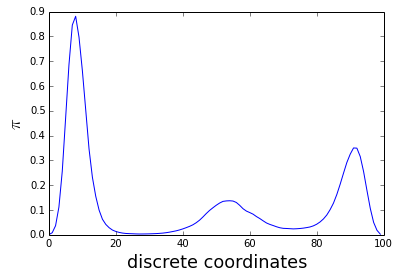
We can then derived the free energy profile (F) or Potential of Mean force (PMF):
and compare to the initial one, used in the toy model:
plot(-log(p0), linewidth=2, label='PMF')
plot(x_lin,E(x_lin), linewidth=2, label='Original potential')
ylabel('Potential', fontsize=18)
xlabel('discrete coordinates', fontsize=18)
grid()
tmp = legend()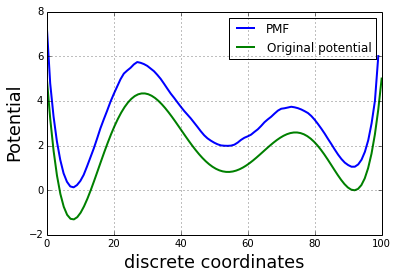
If we normalized with the \(F_{0}\) factor we obtain:
F0 = (log(p0) + E(x_lin)).mean()
plot(F0-log(p0), linewidth=2, label='PMF')
plot(x_lin,E(x_lin), '--', linewidth=2, label='Original potential')
grid()
tmp = legend()